#include <Rcpp.h>
// [[Rcpp::depends(dqrng, BH)]]
#include <boost/random/gamma_distribution.hpp>
#include <dqrng.h>
// [[Rcpp::export]]
Rcpp::NumericVector dqrgamma(std::size_t n, double shape, double scale) {
auto rng = dqrng::random_64bit_accessor{};
auto out = Rcpp::NumericVector(Rcpp::no_init(n));
rng.generate<boost::random::gamma_distribution>(out, shape, scale);
return out;
}Today dqrng version 0.4.0 has been published on CRAN and is now propagating through the mirrors. At the moment we are still working through some breakage with reverse dependencies, but that should clear up soon.
I had already mentioned the changes in a previous post. There is one important change, though: I reverted the breaking change concerning the two argument constructor and seed function from PCG. While it is still true that those have surprising properties, this is not really relevant for the current use-case. Here it is important that the new method dqrng::random_64bit_generator::clone(stream) produces an identical RNG for clone == 0, which can be implemented without influencing the two argument constructor and seed function.
What I like about this release is that different streams of work are coming together quite nicely. For example, Henrik Sloot implemented to ability to access the global RNG within C++ code. In addition, the abstract base class random_64bit_generator now supports methods variate<dist>(param), generate<dist>(container, param) etc. using and inspired by randutils. These make it super easy to create new dqr<dist> functions if needed, e.g.:
Since this new function makes use of the global RNG, it is controlled via the same dqset.seed() as normal functions from the package:
dqset.seed(20240515)
dqrgamma(5, 2, 10)[1] 30.17319 44.74696 12.02194 16.64893 17.34264dqrnorm(5)[1] 0.28924215 -0.87322364 0.91917936 -0.83256856 0.01148021dqset.seed(20240515)
dqrgamma(5, 2, 10)[1] 30.17319 44.74696 12.02194 16.64893 17.34264dqrnorm(5)[1] 0.28924215 -0.87322364 0.91917936 -0.83256856 0.01148021It should not come as a surprise that boost::random::gamma_distribution() produces similarly distributed random variates as the function from base R:
n <- 1e6
shape <- 2
scale <- 10
data.frame(
gamma_R = rgamma(n, shape, scale = scale),
gamma_dq = dqrgamma(n, shape, scale)) |>
pivot_longer(cols = starts_with("gamma")) |>
ggplot(aes(x = value, fill = name)) + geom_histogram(position = "dodge2")`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.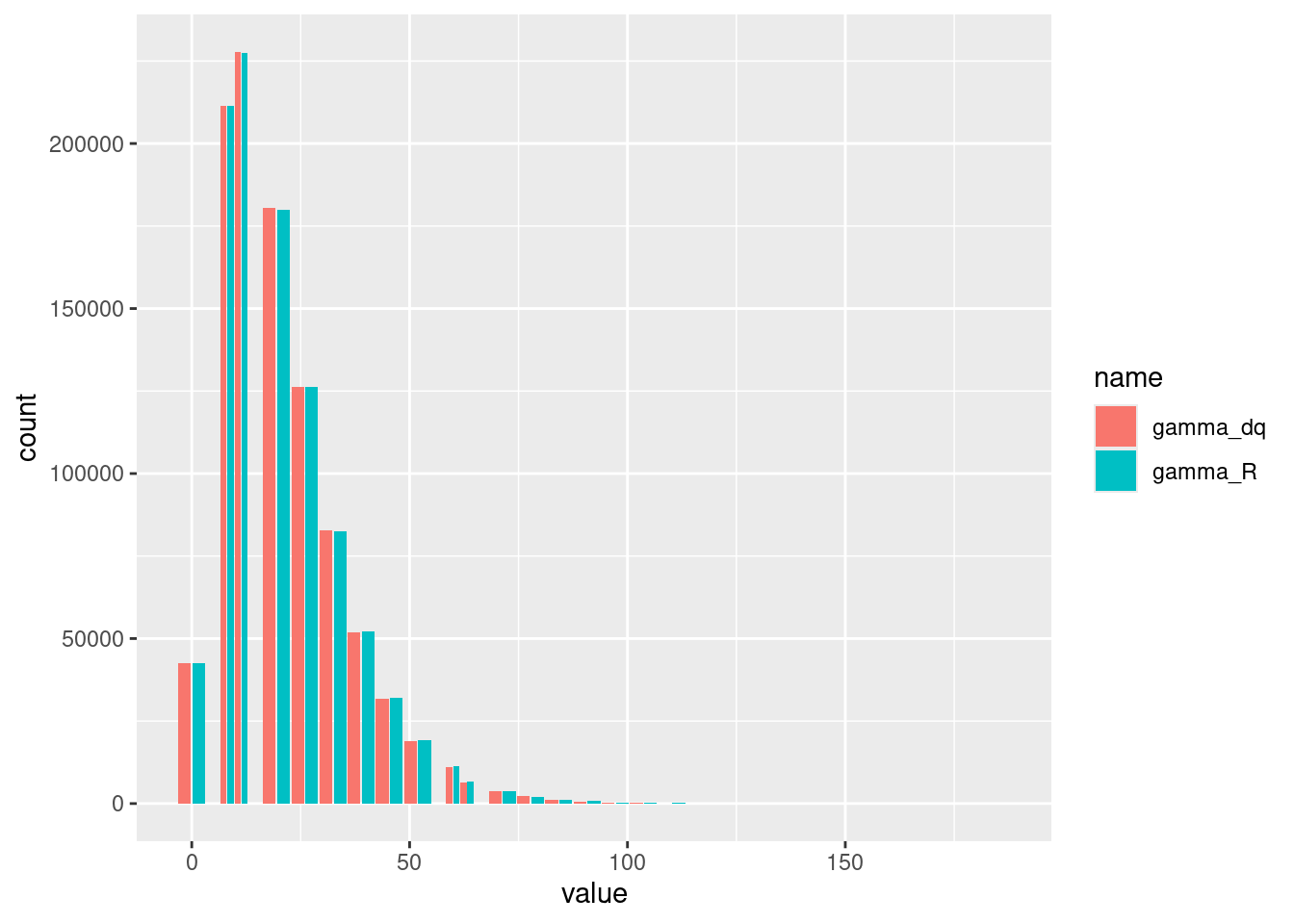
Unfortunately, the Gamma distribution implemented in boost.random is not very fast:
bm <- bench::mark(gamma_R = rgamma(n, shape, scale = scale),
gamma_dq = dqrgamma(n, shape, scale),
check = FALSE)
knitr::kable(bm[, 1:5])| expression | min | median | itr/sec | mem_alloc |
|---|---|---|---|---|
| gamma_R | 133ms | 139ms | 7.162025 | 7.63MB |
| gamma_dq | 159ms | 171ms | 5.814996 | 7.63MB |
It might make sense to implement for example the method from Marsaglia and Tsang (2000).
Alternatively, one can also make use of a feature that I had discussed in another post. It is now possible to register dqrng as user-supplied RNG within R. This way one gets the already fast uniform and normal distribution functions from dqrng together with all the distribution functions available for R. This can make for a very fast combination:
register_methods()
bm <- bench::mark(gamma_R = rgamma(n, shape, scale = scale),
gamma_dq = dqrgamma(n, shape, scale),
check = FALSE)
knitr::kable(bm[, 1:5])| expression | min | median | itr/sec | mem_alloc |
|---|---|---|---|---|
| gamma_R | 96.6ms | 97.7ms | 10.209660 | 7.63MB |
| gamma_dq | 156.8ms | 157.9ms | 6.337992 | 7.63MB |