sample_R <- function (size, prob) {
sample.int(length(prob), size, replace = FALSE, prob)
}Some time ago I learned from a blog post by Thomas Lumley that R uses “sequential sampling” when using weighted sampling without replacement. As a consequence, the probability of an item being included in the sample is not proportional to the item’s weight.
This felt very counter-intuitive at first, but a blog post by Peter Ellis added some further discussion and useful visualizations, which I am adapting here for my needs. For the current use-case we can use a simplified function signature with only the number of selected elements size and the vector of weights prob. A wrapper for base R’s sample would look like:
Adapting the visualization function we get:
library(tidyverse)── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.4 ✔ readr 2.1.5
✔ forcats 1.0.0 ✔ stringr 1.5.1
✔ ggplot2 3.5.1 ✔ tibble 3.2.1
✔ lubridate 1.9.3 ✔ tidyr 1.3.1
✔ purrr 1.0.2
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errorslibrary(glue)
compare_ppswor <- function(n = 10,
N = 20,
reps = 1e5,
prob = 1:N / sum(1:N),
FUN = sample_R,
m = "R's native `sample()` function."){
prob <- prob / sum(prob)
samples <- lapply(rep(n, reps), function(size){
FUN(size = size, prob = prob)
})
p <- tibble(
original = prob,
selected = as.numeric(table(unlist(samples))) / (reps * n)
) |>
ggplot(aes(x = original, y = selected)) +
geom_abline(slope = 1 , intercept = 0, colour = "orange") +
geom_point(colour = "steelblue") +
labs(x = "Original 'probability' or weight",
y = "Actual proportion of selections",
subtitle = glue("Population of {N}, sample size {n}, sampling without replacement.\nUsing {m}"),
title = "Use of `sample()` with unequal probabilities of sampling") +
coord_equal()
p
}
compare_ppswor()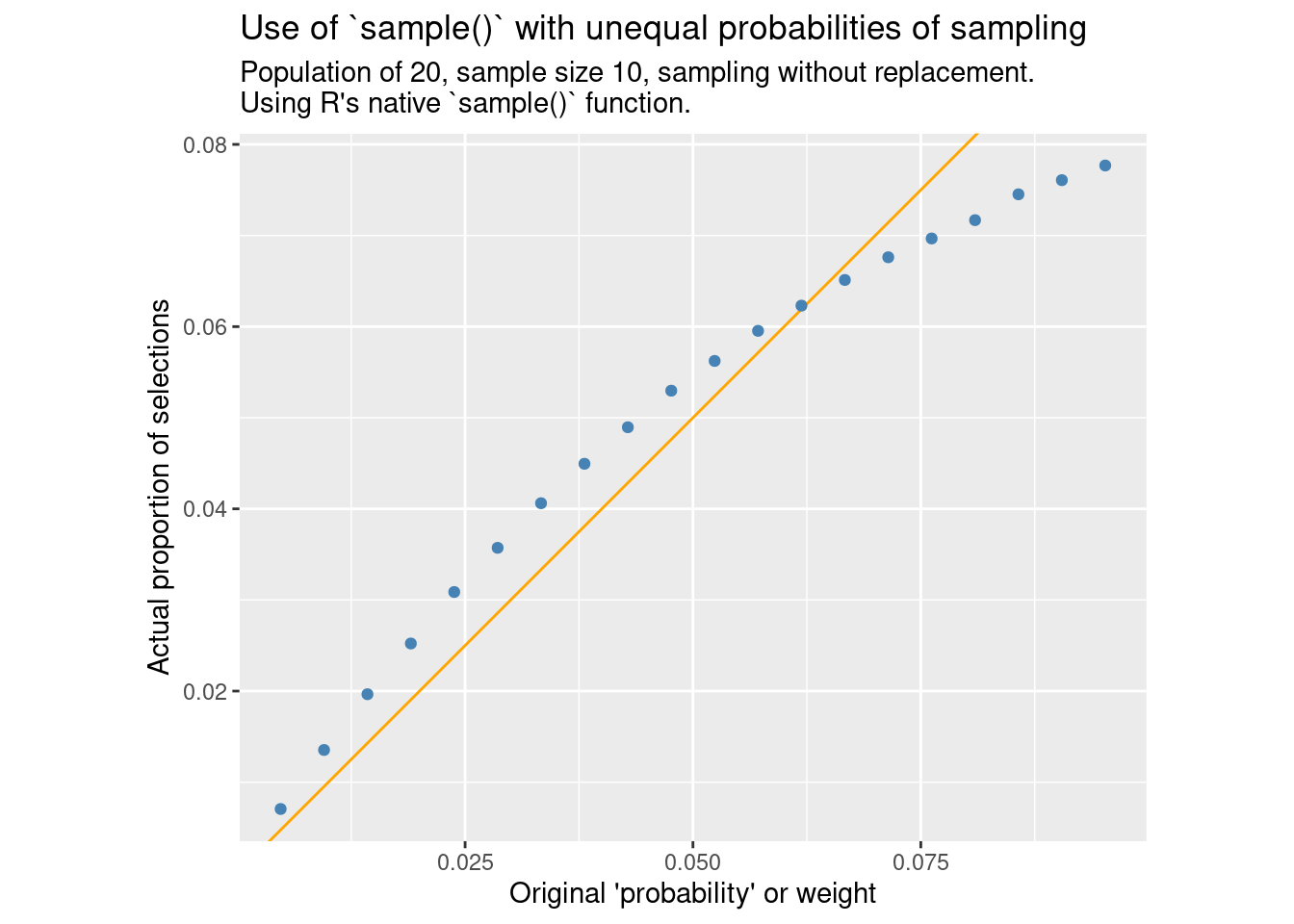
Here we see that the third of the items with the highest weight has a reduced probability of inclusion, while the other two thirds are slightly above the expected probability.
Nice, but the main point of the original blog post was to note that the sampling method suggested by Efraimidis and Spirakis (2006) has the same problem. Using the more stable exponential rank reformulation by Müller (2016, 2020) we divide the original weights by random deviates with exponential distribution. The size largest (or depending on the formulation smallest) values provide the random sample. In C++:
#include <Rcpp.h>
// [[Rcpp::depends(dqrng)]]
#include <dqrng.h>
#include <dqrng_distribution.h>
// [[Rcpp::export]]
Rcpp::IntegerVector sample_exp(int size, Rcpp::NumericVector prob) {
auto rng = dqrng::random_64bit_accessor{};
auto n = prob.length();
Rcpp::IntegerVector index(n);
std::iota(index.begin(), index.end(), 1);
Rcpp::NumericVector weight(n);
rng.generate<dqrng::exponential_distribution>(weight);
weight = - prob / weight;
std::partial_sort(index.begin(), index.begin() + size, index.end(),
[&weight](size_t i1, size_t i2) {return weight[i1-1] < weight[i2-1];});
Rcpp::IntegerVector result(size);
std::copy(index.begin(), index.begin() + size, result.begin());
return result;
}compare_ppswor(FUN = sample_exp, m = "Efraimidis and Spirakis (2006).")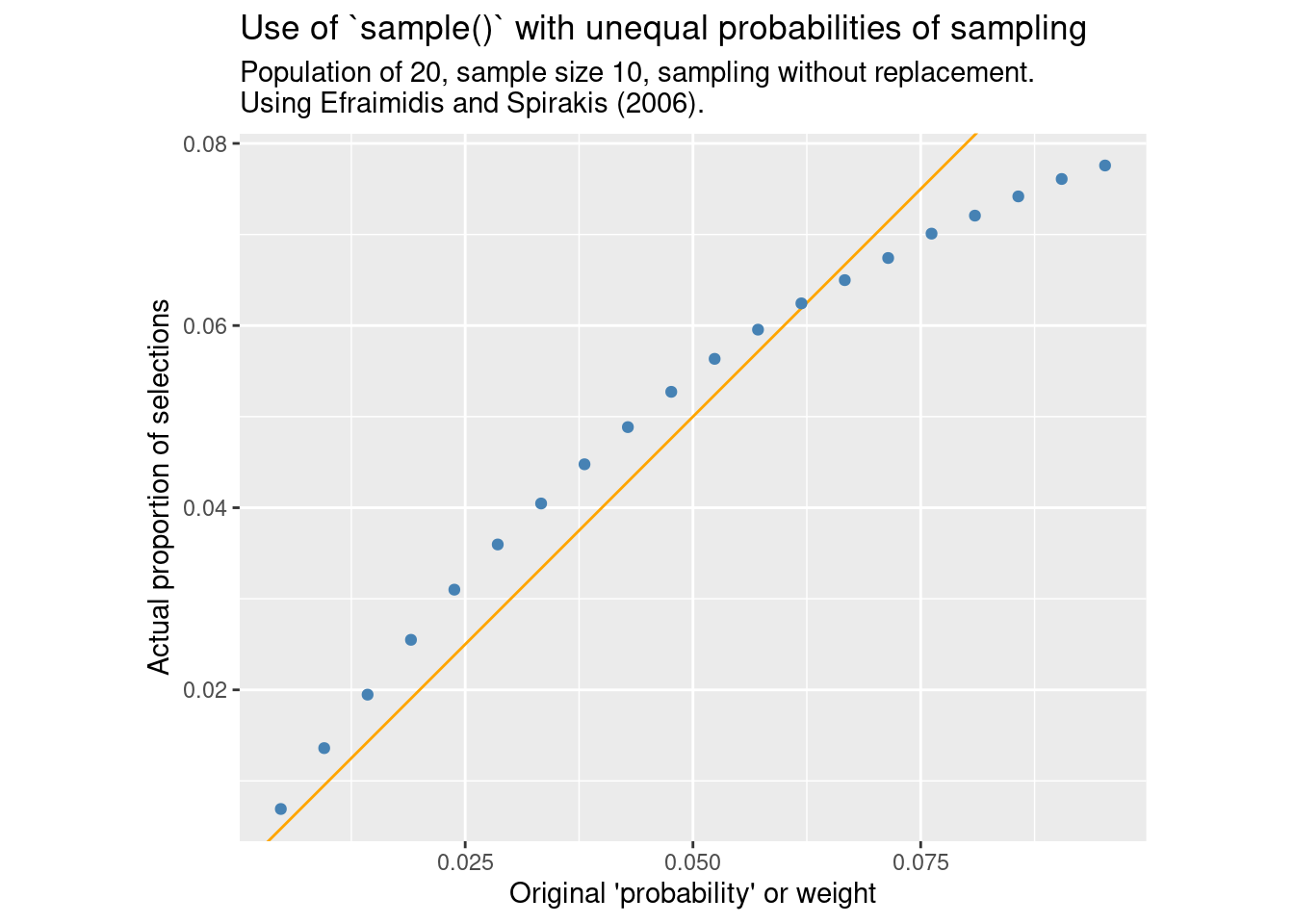
Indeed, the pattern is the same as seen by base R’s sample() in that the probability of inclusion is not proportional to the supplied weight.
The algorithm suggested by Brewer (1975) solves this problem. That paper is a bit hard to get by, but fortunately Tillé and Matei (2023) provide an implementation:
sampling::UPbrewer = function (pik, eps = 1e-06) {
if (any(is.na(pik)))
stop("there are missing values in the pik vector")
n = sum(pik)
n = .as_int(n)
list = pik > eps & pik < 1 - eps
pikb = pik[list]
N = length(pikb)
s = pik
if (N < 1)
stop("the pik vector has all elements outside of the range [eps,1-eps]")
else {
sb = rep(0, N)
n = sum(pikb)
for (i in 1:n) {
a = sum(pikb * sb)
1 p = (1 - sb) * pikb * ((n - a) - pikb)/((n - a) - pikb * (n - i + 1))
p = p/sum(p)
p = cumsum(p)
u = runif(1)
for (j in 1:length(p)) if (u < p[j])
break
sb[j] = 1
}
s[list] = sb
}
s
}- 1
-
Replace this by
p = (1 -sb) * pkibto get “sequential sampling”.
Ignoring the handling of very small and large inclusion probabilities for now we can translate this into C++:
#include <Rcpp.h>
// [[Rcpp::depends(dqrng)]]
#include <dqrng.h>
// [[Rcpp::export]]
Rcpp::IntegerVector sample_brewer(int size, Rcpp::NumericVector prob) {
auto rng = dqrng::random_64bit_accessor{};
Rcpp::NumericVector pikb = size * prob / Rcpp::sum(prob);
auto N = pikb.length();
Rcpp::NumericVector mask(N);
Rcpp::NumericVector p(N);
Rcpp::IntegerVector result(size);
for (int i = 0; i < size; ++i) {
double a = Rcpp::sum(pikb * mask);
p = (1 - mask) * pikb * ((size - a) - pikb) / ((size - a) - pikb * (size - i));
p = p / Rcpp::sum(p);
p = Rcpp::cumsum(p).get();
double u = rng.uniform01();
int j = std::upper_bound (p.begin(), p.end(), u) - p.begin();
mask[j] = 1;
result[i] = j + 1;
}
return result;
}compare_ppswor(FUN = sample_brewer, m = "Brewer (1975) as implemented by Tillé/Matei.")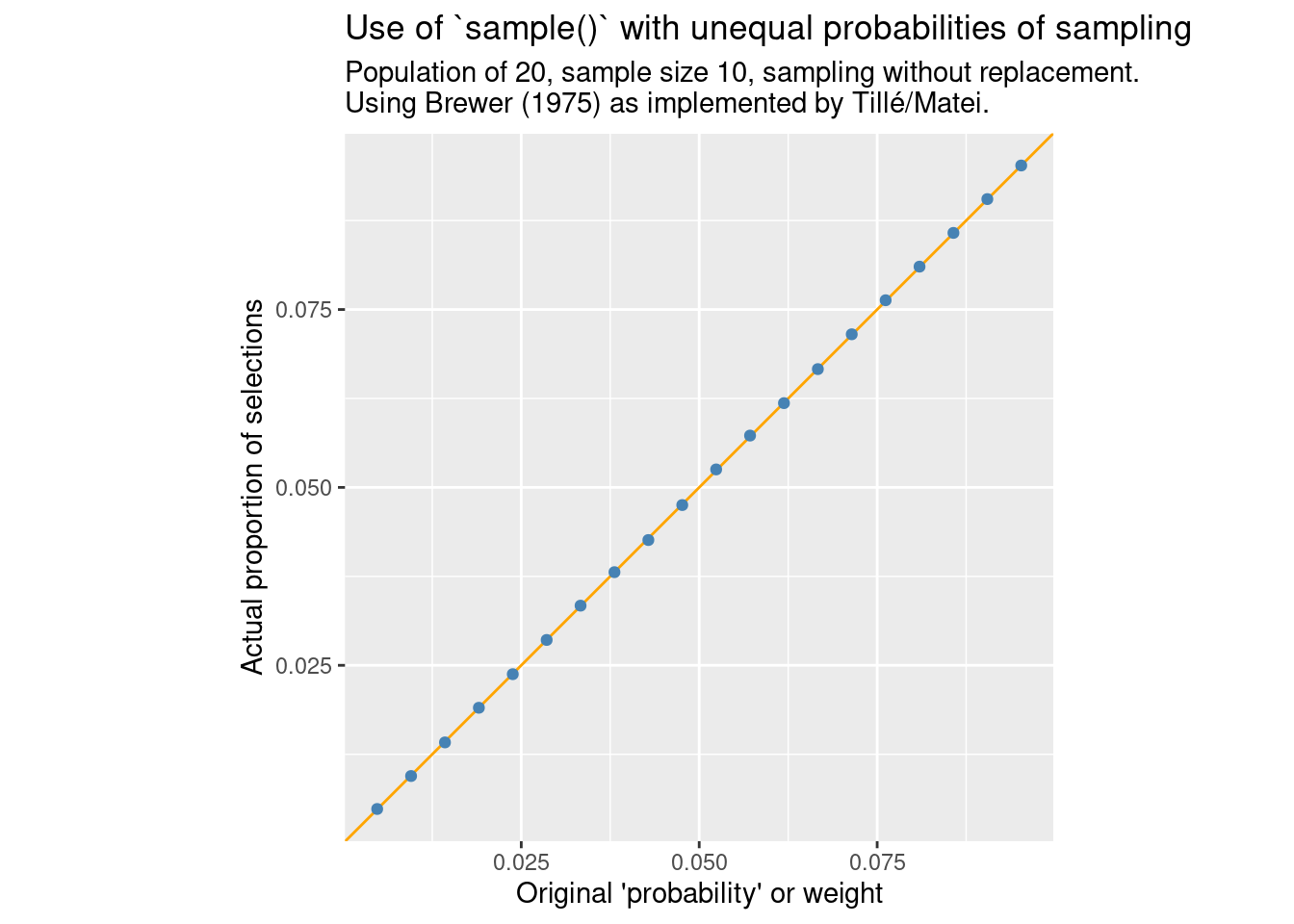
Here we get the expected proportionality between the probability of inclusion and original weight. Of course, one reason to use Efraimidis and Spirakis (2006) is that this method has the potential of being much faster. So how does performance compare here?
bp1 <- bench::press(
n = 10^(1:4),
r = c(0.5, 10^-(1:3)),
{
size <- ceiling(r * n)
prob <- sample(n)
bench::mark(
sample_R(size, prob),
sample_brewer(size, prob),
sample_exp(size, prob),
check = FALSE,
time_unit = "s"
)
}
) |> mutate(label = as.factor(attr(expression, "description")))Warning: Some expressions had a GC in every iteration; so filtering is
disabled.
Warning: Some expressions had a GC in every iteration; so filtering is
disabled.ggplot(bp1, aes(x = n, y = median, color = label)) +
geom_line() + scale_x_log10() + scale_y_log10() + facet_wrap(vars(r))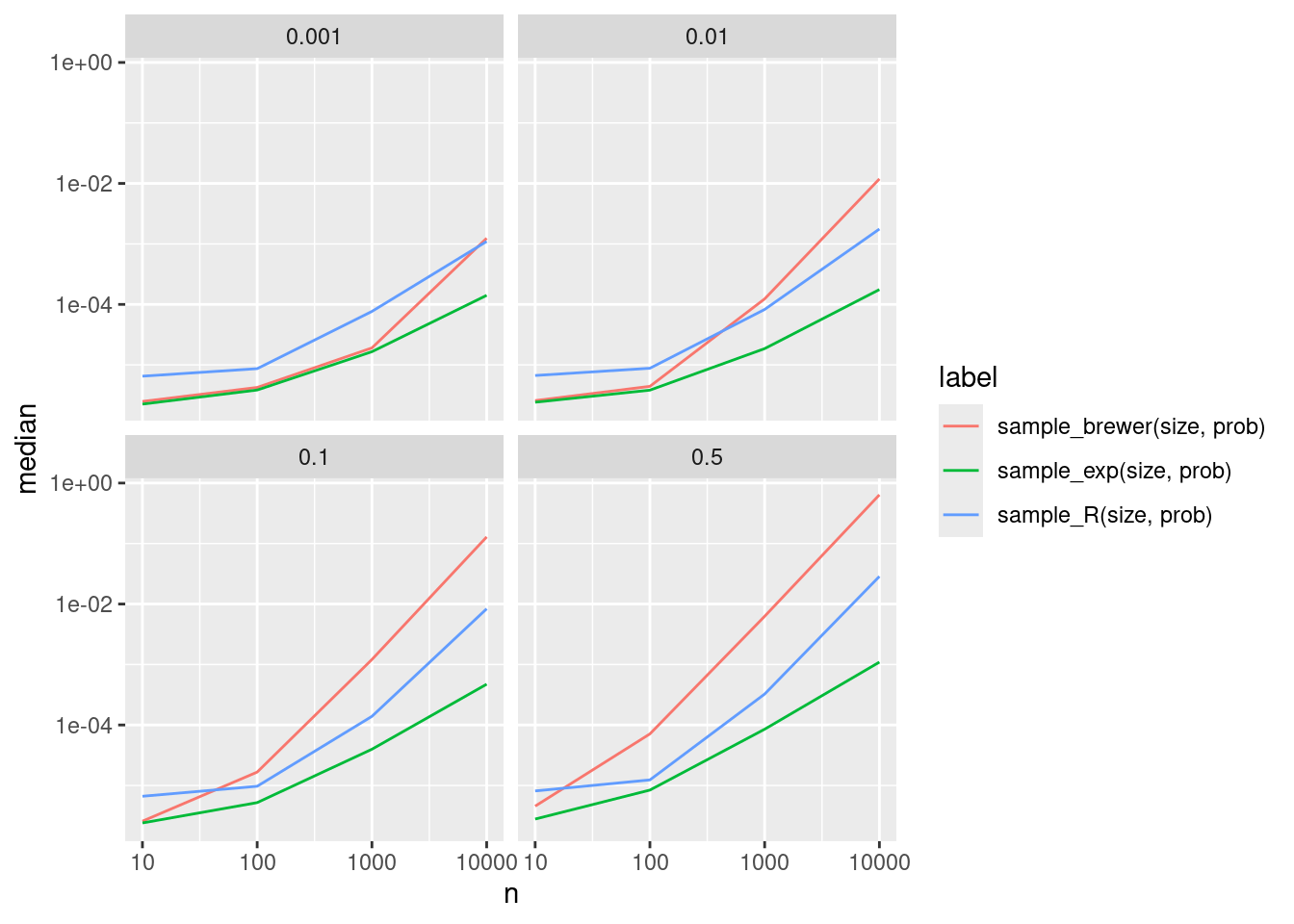
Now this does not look good. In many cases Brewer’s method is even slower than the already quite slow default method from R, while the exponential rank method is quite a bit faster.
Fortunately, it should be OK to use “sequential sampling” if the ratio of selected elements is low enough:
compare_ppswor(N = 100, FUN = sample_exp, m = "Efraimidis and Spirakis (2006).")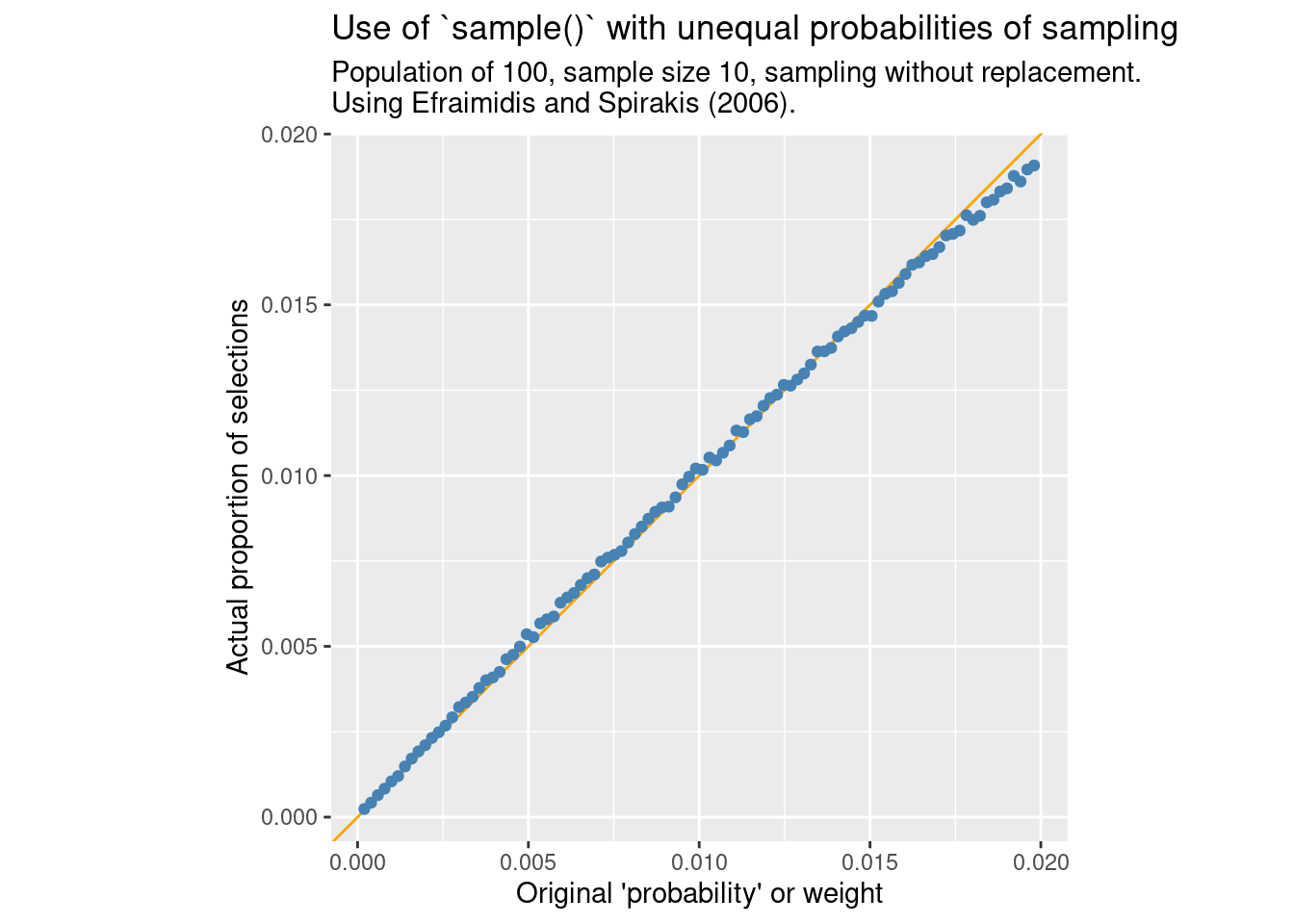
But for higher selection ratios offering Brewer’s algorithm at least as an option seems to be prudent before including this in dqrng. More to come …